Facilities
The Genomics Unit
The Genomics Unit (GU) provides research groups with technical support for genomic applications (routine use and development of tailor-made protocols), accommodates any research need (e.g. automation of library preparation, protocol optimization), implements and applies the latest technologies and protocols. It is managed in collaboration between the IIT CGS and the Department of Experimental Oncology of the European Institute of Oncology of IEO. The GU is equipped with the most advanced genomic technologies, including 3 next generation sequencing platforms (Novaseq 6000, HiSeq 2000 and MiSeq - Illumina), single cell genomics (10X Chromium, Chromium Connect), direct sequencing (GridIon - Oxford Nanopore) and various gene expression analysis platforms (nCounter - Nanostring). The unit is also equipped with liquid handling robots for automation and micromanipulation (Beckman, Cellink) and various instruments for the quantification and quality control of DNA and RNA (Bioanalyzer, LabChipGX and Tape Station). The GU offers the following services:
DNA Analysis
- Whole Genome Analysis (WGA) - Resequencing, De Novo
- Targeted DNA Analysis (Exome sequencing, Custom enrichment)
- Chromatin (ChIP-Seq)
- Methylation analysis (RRBS, Whole Genome methylation)
RNA Analysis
- Expression Profiling (Whole Transcriptome)
- Small RNA sequencing (sRNA-seq)
- Targeted RNA analysis (nascent RNA analysis (4sU/TT-seq), Methylated RNA analysis, immunoprecipated RNA (RIP/RAP))
Other services
- ChIPSeq Library Generation(Beckman Automation), Target Enrichment
- Quality Control (QC) - DNA and RNA
Single-cell
- Single Cell Gene Expression
- Single Cell Immune Profiling
- Single Cell CNV
- SIngle Cell ATAC
The Computational Unit
The Computational Unit (CU) manages the computational framework and pipelines associated with all types of sequencing applications. The computational workflows fully support the activity of the Genomics Unit, closely integrated with the activity of the Research Groups. A sample manager developed in-house oversees and keeps track of sequencing requests and results. The latter are automatically visible on HTS-flow (Bianchi et al., 2016), a Workflow Management System that is accessible online and implements primary and secondary data analysis pipelines for all types of sequencing experiments (e.g. ChIP-seq, RNA-seq, and DNA-seq). This asset allows a straight-forward access to genomics data and their analysis, empowering wet-lab scientists with minimal training, and relieving bioinformaticians from performing routine tasks.
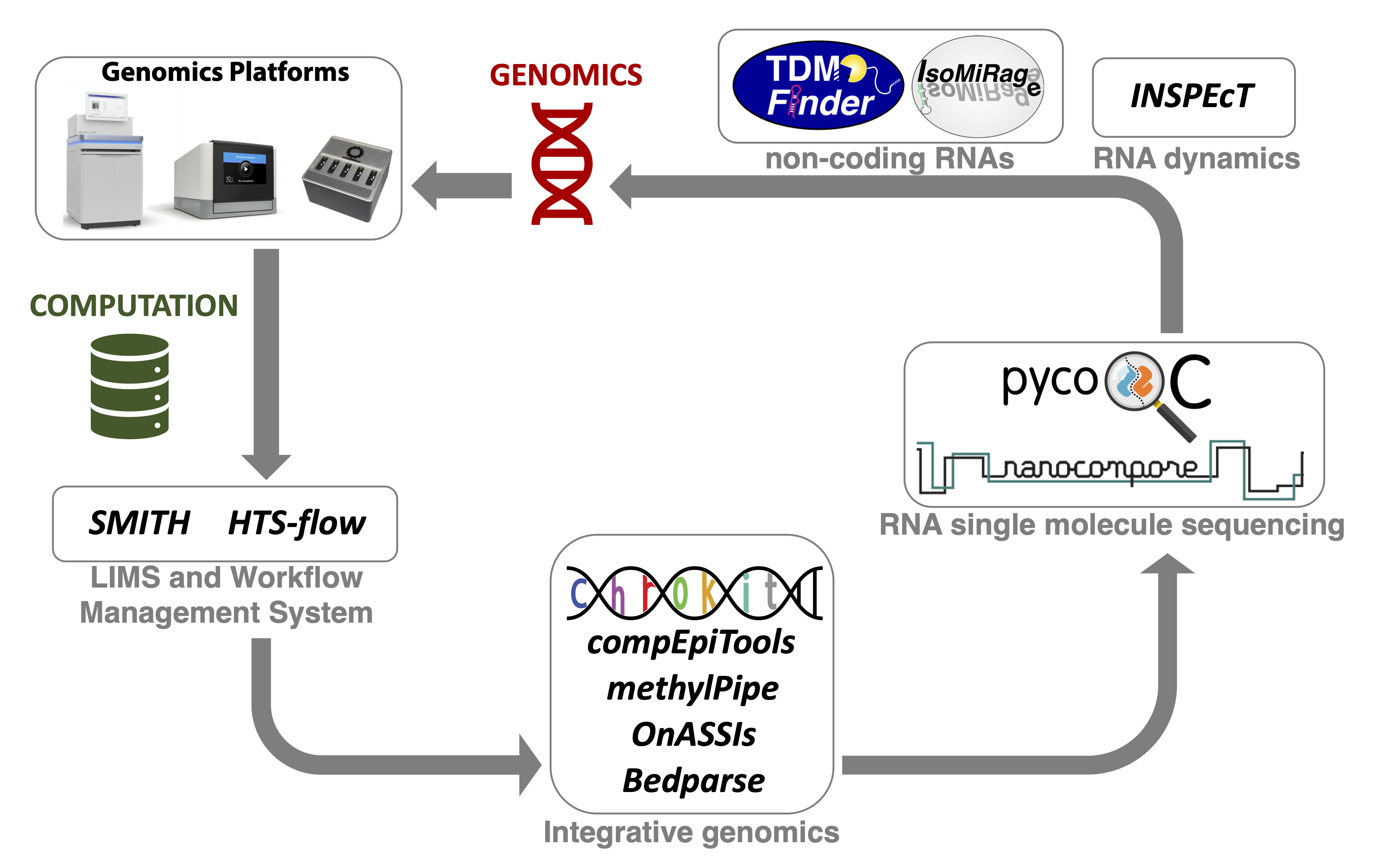
A number of software are being developed by the Research Groups, some of which are also integrated within HTS-flow, including:
- INSPEcT+ (de Pretis et al., 2015) for the quantification of RNA dynamics through the mathematical modelling of total and nascent RNA sequencing data
- INSPEcT- (Furlan et al., 2020) for the quantification of RNA dynamics without requiring the profiling of nascent transcription
- INSPEcT-GUI (de Pretis et al., 2020) for the interactive visualisation and modelling of gene-level RNA dynamics
- compEpiTools and methylPipe (Kishore et al., 2015) for the integrative analysis of epigenomics data, including whole-genome DNA methylation data
- Onassis (Galeota E and Pelizzola M 2020) for the integrative analysis of omics data through their semantic annotation
- IsomiRage (Muller et al., 2014) for the quantification of miRNAs and isoforms expression
- ChroKit (The Chromatin ToolKit from O. Croci) is a Shiny-based framework to interactively analyse and visualize genomic data.
- Nanocompore (Leger et al., 2019) for the detection of RNA modifications by comparative Nanopore direct RNA sequencing