The information encoded in the genome needs to be timely dispatched in response to cellular and organismal needs. This implies a central role of transcription factors and more in general transcriptional regulators in the control of RNA expression in response to external cues. Our laboratory is focusing on the study of transcription factors that control cell growth, cell proliferation and cell identity with the aim of understanding their physiological and pathological function. In particular, we are studying c-Myc, a basic helix-loop-helix transcription factor, which is a general regulator of cell growth and metabolism, and YAP/TAZ, two transcriptional coactivators originally identified as effectors of the Hippo signalling pathway. We make use of sophisticated experimental models for loss of function and gain of function genetic studies combined to genomic analyses based on Next generation sequencing technologies to profile genome wide transcription factors distribution, epigenetic remodeling, chromatin accessibility and 3D nuclear organization. These datasets are integrated with global expression profiling as well as well as single cell mRNA expression in order to (i) uncover relevant transcriptional programs, (ii) define mechanisms for controlling gene expression, (iii) unravel transcription factor networks, and (iv) identify cancer cells liabilities.
YAP/TAZ in tissue homeostasis and cancer
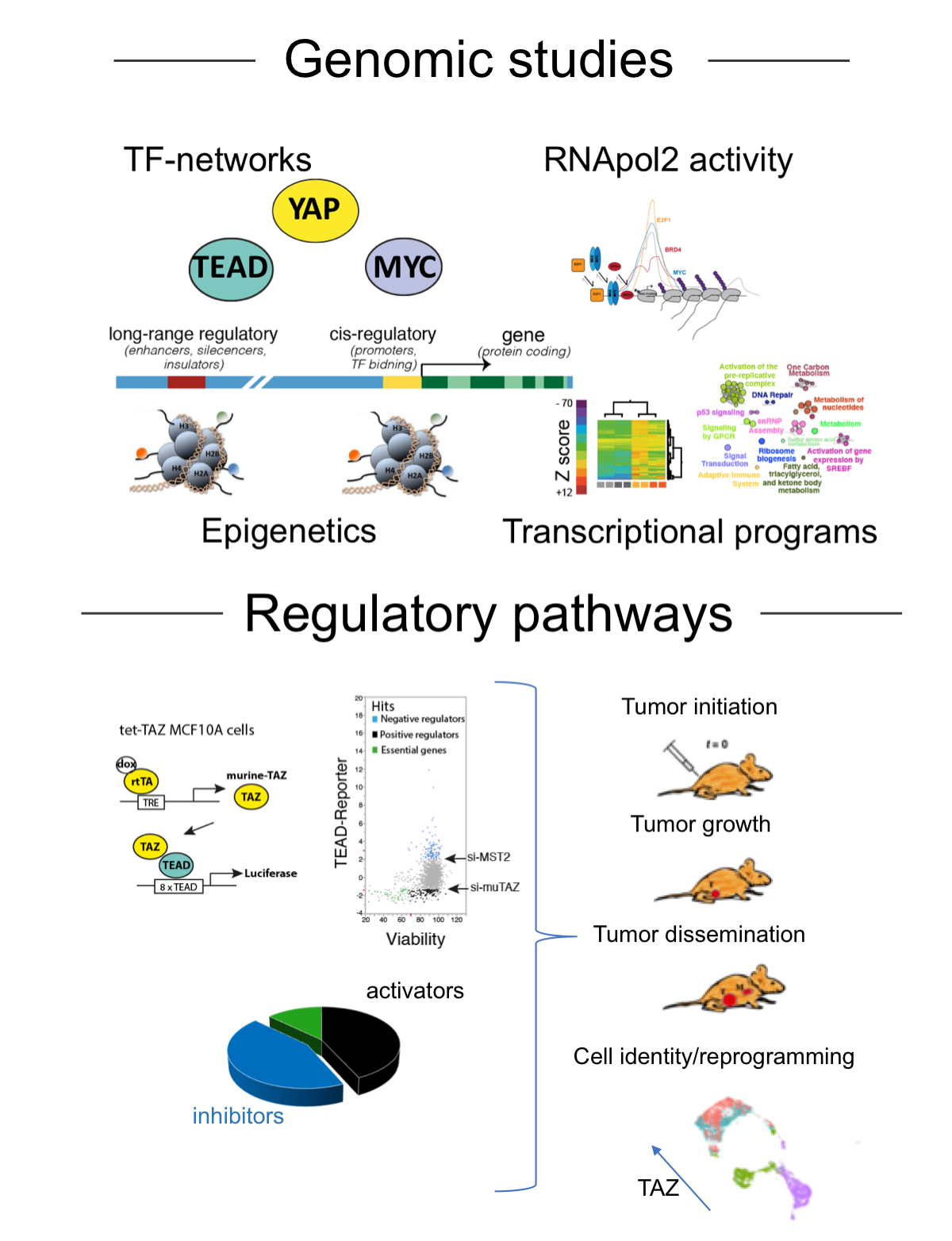
YAP and TAZ are two transcriptional co-activators originally identified as regulated by the Hippo pathway and later recognized as key effectors of cytoskeletal tension, cell polarity, cell-to-cell contacts and G-protein coupled receptors. As such, YAP/TAZ are emerging as essential genes that are able to integrate chemical and mechanical signals in order to regulate tissue growth during development, regeneration and cancer. In several cancer types, YAP/TAZ activity has been linked to mesenchymal-like features, metastatic potential, chemo-resistance and cancer stem cell properties. We have two major research lines:
Genomic study of YAP function in the control and cell identity. In the liver, activation of YAP supports proficient cell proliferation and triggers de-differentiation of hepatocytes towards a bi-potent progenitor-like state. How YAP may control such a large number of cellular processes is still to be fully understood. To address this, we are conducting a genome-wide studies in vivo. This will provide a genome scale understanding of how YAP controls gene expression and will reveal principles in gene regulation which account for YAP ability to orchestrate development, regeneration and cell reprogramming.
Upstream regulation of YAP/TAZ. While YAP/TAZ are frequently stabilized and activated in cancer, pathways leading to their deregulation are yet to be fully identified. To address this, we recently conducted a genetic screen where we identified novel genes that modulate YAP/TAZ activity. We are investigating these genes to gain insights on mechanisms of YAP/TAZ regulation and to assess their functional relevance in cancer development.
Targeting Myc in cancer
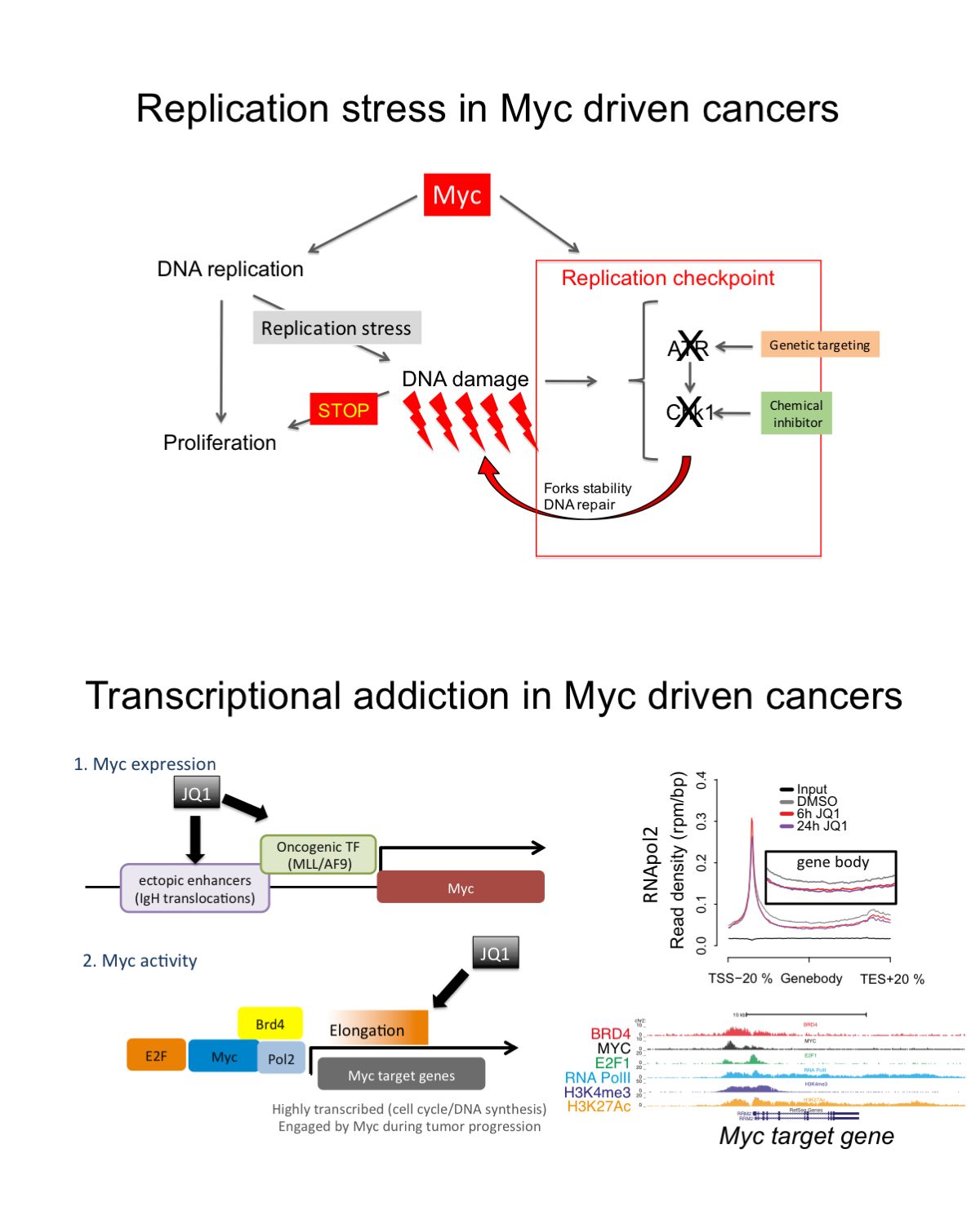
c-Myc (Myc) is an oncogenic transcription factor that, given its essential role in cancer cells, is receiving considerable attention as a therapeutic target. Yet, its biochemical properties hamper its direct inhibition by small molecules. We are circumventing this, by exploiting genetic dependencies in Myc-driven cancers. We are focusing on two aspects:
Therapeutic induction of replicative stress. This research line stems from our early discovery that Myc, while promoting DNA replication, also restrains inherent replicative stress by actively engaging “safeguarding pathways”. This renders Myc-driven tumors liable to undergo deadly replicative stress responses when such mechanisms are disengaged. Based on this rationale, we are conducting genetic screens to identify genes essential for genome stability in Myc over-expressing cells. We are now focusing on transcriptional regulators and on their role in the maintenance of genome stability during DNA replication.
Transcriptional addiction in cancer cells. Oncogenic Myc controls the expression of a thousand transcripts linked to cell cycle regulation and cellular metabolism, thus promoting tumor cells proliferation and fitness. This widespread transcriptional control leads to oncogenic addiction and implies that the selective pharmacological targeting of Myc transcriptional activity would represent an effective avenue to decommission Myc in cancer cells.
The rapid raise and use of genome-wide techniques raises the need for the development of computational and bioinformatics tools to integrated and comparative analysis of genomic data. To this end, we have developed two computational tools, TFARM and ChroKit, designed for the integrated analysis of chromatin modifications and transcription factors binding. TFARM is a suite that uses novel algorithms based on association rules and ranking metrics driven by the definition of an Importance index, to infer functional and physical association of transcription factors, thus providing information on transcription factors networks and transcription factories. Chrokit is multiplatform software with an advanced graphical interface which provides intuitive and efficient tools for the analysis of genomic data from ChIP-seq, RNA-seq and other similar techniques.
1. Rohban S, Campaner S. Myc induced replicative stress response: How to cope with it and exploit it. Biochim Biophys Acta. 2015 May; 1849 (5):517-24. doi: 10.1016/j.bbagrm.2014.04.008.
2. Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, Tonelli C, Fagà G, Bianchi V, Ronchi A, Low D,
Müller H, Guccione E, Campaner S, Amati B. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014 Jul 24;511(7510):488-92. doi: 10.1038/nature13537.
3. Murga M*, Campaner S*, Lopez-Contreras AJ, Toledo LI, Soria R, Montaña MF, D'Artista L, Schleker T, Guerra C, Garcia E, Barbacid M, Hidalgo M, Amati B,
Fernandez-Capetillo O. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011 Nov 27; 18 (12):1331-5. doi: 10.1038/nsmb.2189.
4. Campaner S, Spreafico F, Burgold T, Doni M, Rosato U, Amati B, Testa G. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol Cell. 2011 Aug 19;43(4):681-8. Doi: 10.1016/j.molcel.2011.08.007.
5. Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjö S, Murga M, Fernandez-Capetillo O, Barbacid M, Larsson LG, Amati B. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat. Cell Biol. 2010 Jan; 12 (1):54-9; sup pp 1-14. doi: 10.1038/ncb2004.
