Genomic Science
The scientific mission of the Research Line in Genomic Science is the development and application of advanced genomics technologies for the discovery of the molecular mechanisms underlying human disease, with a particular focus on cancer.
We study the RNA regulatory mechanisms that control gene expression at transcriptional, post-transcriptional and epigenetic level, and aim at identifying disease markers or potential targets for pharmacological intervention.
Main Scientific Aims
At the CGS, experimental research and computational approaches are integrated within and across the two Experimental (Non-coding Genome and Cancer Biology) and two Computational (Epigenomics and Transcriptional Regulation and Computational RNA Biology) Research Groups. A pivotal role is played by our first-rate Technological Facilities (Genomics Unit and Computational Unit), state-of-the-art methodologies and analytical tools, sophisticated experimental models and easy access to clinical samples.
Our long-term goals are:
- Developing applications of Next-generation Genomics Technologies
- Strengthening IIT’s scientific leadership in the fields of RNA biology, RNA technology and Cancer
ncRNAs in cancer
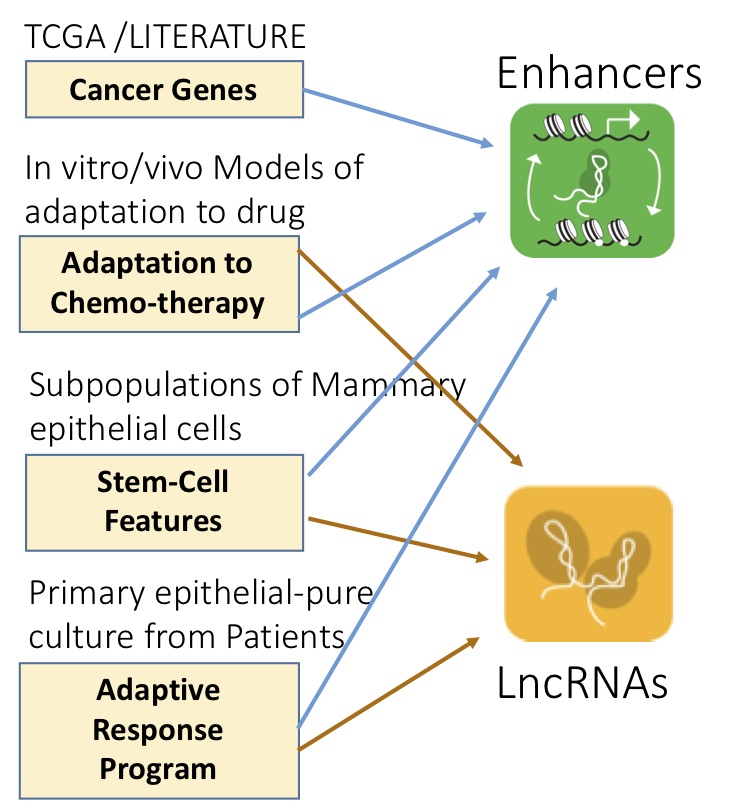
The eukaryotic genome contains non-coding elements that contribute, both at RNA (lncRNAs, miRNAs) and DNA level (enhancers), to shaping the cell transcriptome in both physiology and disease. We aim at understanding the mechanisms and dynamics regulating transcriptional and regulatory networks relevant to cancer, focusing, in particular, on Cancer Evolution and on the Adaptive Response (transcriptional-epigenetic) pathways of Chemo-Adaptation and Therapy resistance.
Impact on human disease: A promising strategy for halting cancer evolution consists in identifying and targeting any of the bottlenecks that characterize the dynamic processes of adaptation of cancer cells. As they play a role in epigenetic and transcriptional dynamics and act as branching points with wide-ranging effects, ncRNAs appear to be key targets in adaptation mechanisms and transcriptional reprogramming. Their identification would be a milestone in the understanding of the genomic mechanisms underlying human development and pathologies. A targeted therapy based on these regulators should be successful in eradicating cancer cells surviving the currently available therapies.
RNA modifications in physiology and disease biology

Within the cell, RNA molecules undergo a number of modifications that markedly affect their fate by impacting on all steps of the RNA life cycle (e.g. transcription, processing, stability, translation and folding). We aim at clarifying the fundamental properties of RNA modifications, such as their distribution, molecular function and role in biological processes using tools from both experimental and computational biology.
Impact on human disease: the role of RNA modification pathways in human diseases, such as cancer, developmental disorders or viral infections lies largely unexplored. The characterization of the fundamental properties of RNA modifications is potentially significant in terms of clinical applications and commercial exploitation as it may lead to innovative diagnostic or therapeutic strategies.
Transcriptional and post-transcriptional control mechanisms in cancer
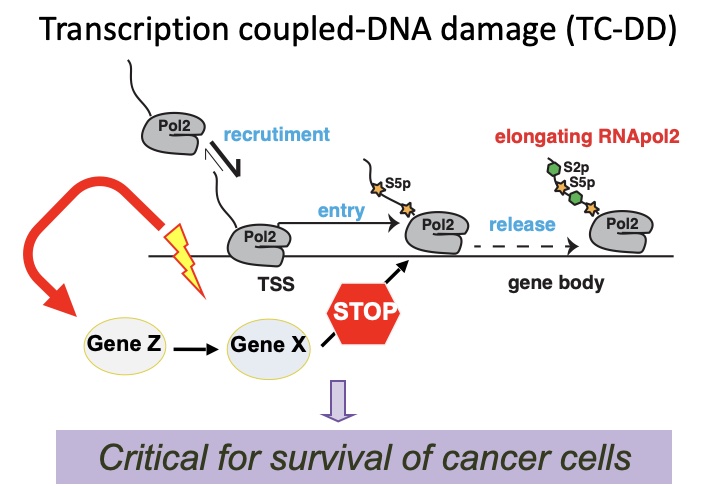
Cancer evolution is driven by the selection of genetic, epigenetic and transcriptional alterations that support oncogenic programs, disengage tumor suppressive pathways, favor adaptation to the tumor microenvironment and foster resistance to therapies. We are studying transcriptional and post-transcriptional regulatory programs in order to understand the mechanisms controlling these key properties of cancer cells. We exploit the latest genomic technologies at population (including RNA-seq, ATAC-seq, 4-SU-seq, and TIDE-seq), single cell (scRNA-seq, scATAC-seq) and single molecule level (Nanopore). Reverse genetics studies, based on gene silencing and genome editing (i.e. shRNA or CRISPR libraries), are used for functional and preclinical validation. We aim at identifying key transcription factors and their essential effectors, unraveling the mechanisms controlling aberrant transcription in cancer cells (transcriptional addiction, enhancers hijacking, alternative splicing, dynamics of RNA processing and decay) and defining cancer cells vulnerabilities (therapeutic targets).
Impact on human disease: a detailed knowledge of how cancer cells process genetic information will instruct the development of targeted therapeutics and new diagnostics (molecular signatures and biomarkers).
Research groups
Non-coding Genome - Francesco Nicassio
Studying the function and regulation of non-coding transcripts (microRNAs and long non-coding RNAs) with a particular emphasis on how regulatory RNAs shape the identity and properties of mammalian cells in cancer stem cells and during cancer evolution (therapy resistance, metastatic spread)
Cancer Biology - Stefano Campaner
Investigating transcriptional and epigenomic alterations in cancer etiology, to uncover regulatory networks causally implicated in oncogenesis and identify cancer cell liabilities
Epigenomics and Transcriptional Regulation - Mattia Pelizzola
Applying an interdisciplinary approach that combines mathematical modeling with genomics to the study of the dynamics of transcription (including RNA synthesis, processing and degradation), and of how these are affected by RNA modifications and chromatin regulation in cancer
Computational RNA biology - Tommaso Leonardi
Developing computational methods and algorithms to study function and regulation of RNA molecules, with a particular focus on the use of direct RNA sequencing approaches (dsRNAseq) based on Nanopore sequencing